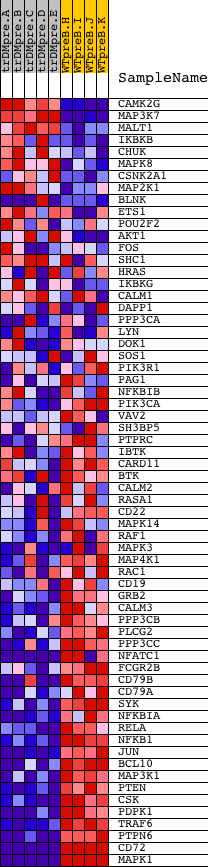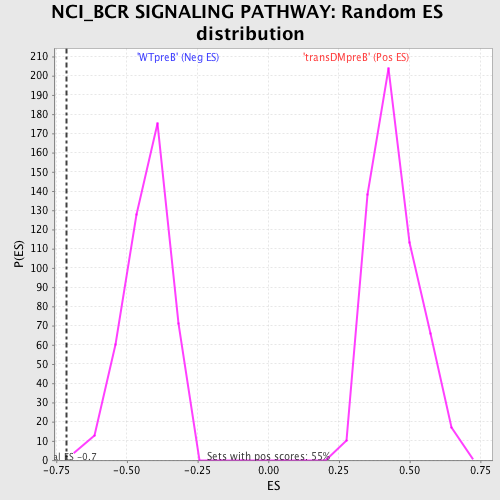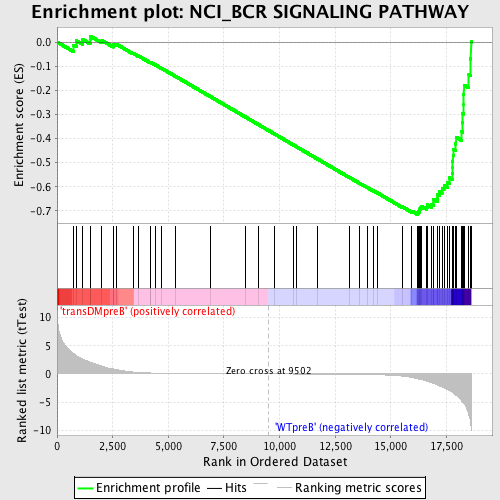
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | NCI_BCR SIGNALING PATHWAY |
| Enrichment Score (ES) | -0.71491987 |
| Normalized Enrichment Score (NES) | -1.6626188 |
| Nominal p-value | 0.0044345898 |
| FDR q-value | 0.031305615 |
| FWER p-Value | 0.226 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CAMK2G | 21905 | 744 | 3.579 | -0.0126 | No | ||
| 2 | MAP3K7 | 16255 | 878 | 3.198 | 0.0049 | No | ||
| 3 | MALT1 | 6274 | 1143 | 2.647 | 0.0110 | No | ||
| 4 | IKBKB | 4907 | 1483 | 2.082 | 0.0087 | No | ||
| 5 | CHUK | 23665 | 1487 | 2.079 | 0.0246 | No | ||
| 6 | MAPK8 | 6459 | 2014 | 1.381 | 0.0069 | No | ||
| 7 | CSNK2A1 | 14797 | 2525 | 0.884 | -0.0138 | No | ||
| 8 | MAP2K1 | 19082 | 2547 | 0.862 | -0.0083 | No | ||
| 9 | BLNK | 23681 3691 | 2670 | 0.771 | -0.0090 | No | ||
| 10 | ETS1 | 10715 6230 3135 | 3419 | 0.324 | -0.0468 | No | ||
| 11 | POU2F2 | 17929 | 3654 | 0.260 | -0.0574 | No | ||
| 12 | AKT1 | 8568 | 4184 | 0.164 | -0.0847 | No | ||
| 13 | FOS | 21202 | 4219 | 0.162 | -0.0853 | No | ||
| 14 | SHC1 | 9813 9812 5430 | 4421 | 0.139 | -0.0950 | No | ||
| 15 | HRAS | 4868 | 4707 | 0.115 | -0.1095 | No | ||
| 16 | IKBKG | 2570 2562 4908 | 5302 | 0.081 | -0.1409 | No | ||
| 17 | CALM1 | 21184 | 6893 | 0.037 | -0.2263 | No | ||
| 18 | DAPP1 | 11159 6445 6446 | 8472 | 0.013 | -0.3113 | No | ||
| 19 | PPP3CA | 1863 5284 | 9030 | 0.006 | -0.3412 | No | ||
| 20 | LYN | 16281 | 9750 | -0.003 | -0.3800 | No | ||
| 21 | DOK1 | 17104 1018 1177 | 10626 | -0.015 | -0.4270 | No | ||
| 22 | SOS1 | 5476 | 10751 | -0.016 | -0.4336 | No | ||
| 23 | PIK3R1 | 3170 | 11694 | -0.031 | -0.4841 | No | ||
| 24 | PAG1 | 15376 | 13138 | -0.066 | -0.5614 | No | ||
| 25 | NFKBIB | 17906 | 13596 | -0.086 | -0.5854 | No | ||
| 26 | PIK3CA | 9562 | 13936 | -0.107 | -0.6028 | No | ||
| 27 | VAV2 | 5848 2670 | 14239 | -0.130 | -0.6181 | No | ||
| 28 | SH3BP5 | 6278 | 14402 | -0.145 | -0.6257 | No | ||
| 29 | PTPRC | 5327 9662 | 15502 | -0.380 | -0.6820 | No | ||
| 30 | IBTK | 19047 | 15935 | -0.604 | -0.7006 | No | ||
| 31 | CARD11 | 8436 | 16201 | -0.863 | -0.7083 | Yes | ||
| 32 | BTK | 24061 | 16235 | -0.895 | -0.7032 | Yes | ||
| 33 | CALM2 | 8681 | 16272 | -0.940 | -0.6979 | Yes | ||
| 34 | RASA1 | 10174 | 16303 | -0.967 | -0.6921 | Yes | ||
| 35 | CD22 | 17881 | 16316 | -0.978 | -0.6852 | Yes | ||
| 36 | MAPK14 | 23313 | 16395 | -1.021 | -0.6815 | Yes | ||
| 37 | RAF1 | 17035 | 16625 | -1.259 | -0.6842 | Yes | ||
| 38 | MAPK3 | 6458 11170 | 16645 | -1.295 | -0.6752 | Yes | ||
| 39 | MAP4K1 | 18313 | 16826 | -1.546 | -0.6731 | Yes | ||
| 40 | RAC1 | 16302 | 16919 | -1.693 | -0.6650 | Yes | ||
| 41 | CD19 | 17640 | 16921 | -1.697 | -0.6520 | Yes | ||
| 42 | GRB2 | 20149 | 17099 | -1.953 | -0.6465 | Yes | ||
| 43 | CALM3 | 8682 | 17112 | -1.978 | -0.6319 | Yes | ||
| 44 | PPP3CB | 5285 | 17175 | -2.093 | -0.6192 | Yes | ||
| 45 | PLCG2 | 18453 | 17303 | -2.308 | -0.6083 | Yes | ||
| 46 | PPP3CC | 21763 | 17392 | -2.468 | -0.5940 | Yes | ||
| 47 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 17547 | -2.734 | -0.5813 | Yes | ||
| 48 | FCGR2B | 8959 | 17622 | -2.880 | -0.5631 | Yes | ||
| 49 | CD79B | 20185 1309 | 17759 | -3.226 | -0.5456 | Yes | ||
| 50 | CD79A | 18342 | 17781 | -3.312 | -0.5213 | Yes | ||
| 51 | SYK | 21636 | 17785 | -3.326 | -0.4959 | Yes | ||
| 52 | NFKBIA | 21065 | 17794 | -3.356 | -0.4705 | Yes | ||
| 53 | RELA | 23783 | 17808 | -3.389 | -0.4451 | Yes | ||
| 54 | NFKB1 | 15160 | 17907 | -3.696 | -0.4219 | Yes | ||
| 55 | JUN | 15832 | 17929 | -3.751 | -0.3942 | Yes | ||
| 56 | BCL10 | 15397 | 18173 | -4.717 | -0.3710 | Yes | ||
| 57 | MAP3K1 | 21348 | 18225 | -4.997 | -0.3353 | Yes | ||
| 58 | PTEN | 5305 | 18232 | -5.045 | -0.2969 | Yes | ||
| 59 | CSK | 8805 | 18265 | -5.201 | -0.2586 | Yes | ||
| 60 | PDPK1 | 23097 | 18270 | -5.228 | -0.2186 | Yes | ||
| 61 | TRAF6 | 5797 14940 | 18289 | -5.308 | -0.1787 | Yes | ||
| 62 | PTPN6 | 17002 | 18509 | -7.257 | -0.1347 | Yes | ||
| 63 | CD72 | 8718 | 18591 | -8.983 | -0.0699 | Yes | ||
| 64 | MAPK1 | 1642 11167 | 18603 | -9.259 | 0.0007 | Yes |